
An ancient bacterium known for devastating and disfiguring its victims has turned to frantically ravaging its own genome to maintain its killer status, according to a new study.
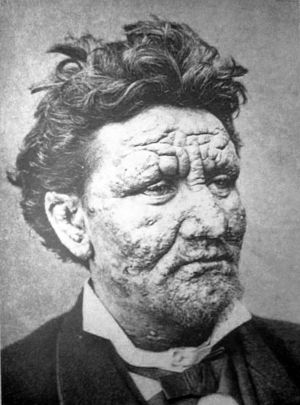
Strains of Mycobacterium leprae—the main bacterium behind leprosy*—are hypermutating and becoming extremely drug resistant. Researchers made the alarming discovery in a survey of 154 M. leprae genomes collected from 25 countries. The survey, published recently in Nature Communications, offers a rare genetic glimpse of the ancient, yet cryptic, bacterium, which still manages to cause 200,000 new cases worldwide each year.
The international team of researchers, led by Stewart Cole of the Ecole Polytechnique Fédérale de Lausanne in Switzerland, noted that the hypermutating state “likely favors the emergence of drug resistance.” But, there’s a catch. Because M. leprae already has a concise genome, it also “could be detrimental and ultimately lethal,” he and his team write. Basically, the revved-up mutation rate could haphazardly damage genes essential for survival.
The genetic peek into what M. leprae strains are up to globally is a rare opportunity, despite the bacterium’s long history with humans. Leprosy likely arose all the way back in the Iron Age (1200-600 BC) and has plagued us ever since. In 1873, physician Gerhard Henrik Armauer Hansen was the first to link bacteria to the disease. He reported that when he dropped water on human cells scraped from a leprous nodule, “rod-shaped bodies” burst out. Those rod-shaped bodies were M. leprae, and the observation was the first time in history that a bacterium was linked to a chronic disease. Hansen’s discovery provided the other name for leprosy, Hansen’s disease.
Perplexing plague
Still, more than a century later, relatively little is known about leprosy. The bacterium is extremely difficult to study because of its unique biology: it grows frustratingly slowly, lives within cells, and transmits cryptically. As such, scientists have yet to figure out how to grow M. leprae alone in labs, how exactly it causes disease, why it is killed by some antibiotics, and how it moves around. It remains a public health threat in South America, Africa, South and Southeast Asia, and Micronesia, where it infects about 200,000 each year.
When it does infect people, researchers know that it usually takes up residence in peripheral nerve cells. Some evidence suggests M. leprae reprograms the cells into a “stem-cell like” state to do its bidding. Infection leads to the inflammation, granulomas, and systemic bacterial spread within the patient. Eventually, patients also suffer sensory loss, disability, and deformations. Left untreated, the infection can be fatal.
So far, researchers still aren’t sure how the bacterium pulls that off or how it arrives in its victims. Direct transmission between people is thought to be the most likely case, but some experts have suggested spread by insects and animals. M. leprae is known to infect mice, armadillos, red squirrels, and some non-human primates.
In the past, the only way researchers could get enough M. leprae for genetic studies was to infect mice and armadillos in lab—then wait a year. M. leprae takes 14 days to go through one generation. By contrast, E. coli can do this in 20 minutes. The slow growth, together with its intracellular residence, make extracting M. leprae from human tissue incredibly difficult.
But, for the new study, Cole and colleagues got around the problem. They worked out and optimized a way to isolate M. leprae from punch biopsies of human tissue. The trick was to disrupt the human cells first, degrade the human DNA, then try to rupture the bacteria and collect as much bacterial DNA as possible.
Deformed DNA
The team analyzed M. leprae genomes from 147 human samples, six red squirrels, and one armadillo from across the globe (see diagram). All samples were from natural infections. The researchers looked at how the strains related to each other, how they appeared to be evolving, and mutations related to drug resistance.
From their relatedness analysis, the researchers found that the strains belonging to the most ancient lineage in their collections were from East Asia. This lines up with previous work suggesting leprosy originated in Eurasia and spread along human migration routes into Africa and the Americas.
But the researchers noted eight strains that were hypermutated, which came from five different subtypes of the bacterium. These hypermutants contained large numbers of mutations throughout their genome. They also all had broken versions of a gene that normally would allow the bacteria to proofread and fix DNA sequence errors, which explains the hypermutation.
-
Geographic distribution of the M. leprae samples used in this study. World map shows the number of registered cases of leprosy per 10,000 population (prevalence rates) in 2015 as reported by the World Health Organization.
-
Timeline of the leprosy treatment and emergence of drug resistance in the XDR strains. Mutated genes conferring resistance to the corresponding drugs are shown in red. Arrows span from the onset of disease to the end of treatment. Horizontal lines show the period when a drug was given. Dotted lines mean irregular treatment. CAM chloramphenicol, CLO clofazimine, DDS dapsone, DPT thiambutosine (diphenylthiourea), ETO ethionamide, INH isoniazid, KAN kanamycin, LVX levofloxacin, MIN minocycline, OFX ofloxacin, PTO protionamide, RIF rifampicin, SMP sulfamethoxypyridazine, SPX sparfloxacin, STR streptomycin, and TZA thiozamin.
The team also noted step-wise development of antibiotic resistance, particularly in some of the hypermutants. Since the 1980s, leprosy has been treated with a combination of two to three antibiotics, typically rifampicin, dapsone, and clofazimine, though it’s unclear how clofazimine kills M. leprae. Prior to that, doctors sometimes prescribed single antibiotics.
For several strains that were resistant to three or more drugs (extensively drug resistant or XDR strains), the researchers looked back at the medical records of the patients from which doctors isolated the strains. The researchers noted that, in several cases, the XDR strains infected patients over decades, with resistance to individual drugs developing one by one as new drugs were tried.
“Drug resistance is alarming for leprosy control,” the authors note. And their new study dug up completely new mutations that may make bacteria resistant to drugs in never-before-seen ways.
“Our discovery of these mutations… should encourage further experimentation in order to establish their true role and contribution to antimicrobial resistance,” the authors conclude.
*A second leprosy bacterium, Mycobacterium lepromatosis, was discovered in 2008 and has been found infecting red squirrels and humans.
Nature Communications, 2018. DOI: 10.1038/s41467-017-02576-z (About DOIs).
https://arstechnica.com/?p=1250927

